Urban wild bees act as 'microbial sensors' of city health
Gut metagenome analysis of solitary mason bees reveals how urban environments shape diet, microbiome stability, pathogen exposure, and antibiotic resistance.
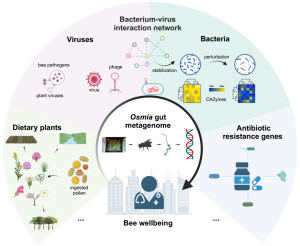
BEIJING, XICHENG, CHINA, December 23, 2025 /EINPresswire.com/ -- As cities grow and natural habitats shrink, urban wildlife must adapt to rapidly changing environments. A new study published in Insect Science shows that the guts of urban-dwelling wild bees contain detailed microbial signatures that reflect both bee health and the quality of the surrounding environment, offering a powerful new tool for monitoring ecological well-being in cities.Researchers at Xi’an Jiaotong-Liverpool University (XJTLU) used metagenomic sequencing of the solitary mason bee Osmia excavata to analyze dietary pollen, gut bacteria and viruses, and antibiotic resistance genes across 10 urban agricultural sites in Suzhou, China. Their findings reveal how gut community data from a single tiny pollinator can expose hidden environmental pressures such as floral scarcity, pathogen spillover, and chemical contamination.
“Our study shows that the gut of a wild bee can act as a sensitive biological sensor of urban environmental quality,” said corresponding author Dr. Min Tang of XJTLU. “By integrating diet, bacteria, viruses, and antibiotic resistance into a single metagenomic workflow, we capture ecological pressures that traditional field surveys often overlook.”
Bee gut DNA reveals constrained diets shaped by urban vegetation
Metagenomic analysis of plant DNA showed that urban Osmia bees rely heavily on a small subset of floral resources, especially Brassica crops and the ornamental tree Platanus. Because Platanus is not typically preferred by bees, its frequent appearance suggests that city bees often forage opportunistically when options are limited.
Diet patterns varied across sites and closely matched local vegetation, demonstrating how the structure of urban landscapes strongly influences seasonal foraging opportunities.
A stable but environmentally sensitive microbiome
Across Suzhou’s varied habitats, the bees maintained a relatively consistent “core” gut microbiome dominated by Gammaproteobacteria, particularly the bacterial genus Sodalis. This symbiont encoded the widest variety of enzymes needed to break down the pollen coat, underscoring its importance for bee nutrition.
At two sites, however, Sodalis was nearly absent replaced by opportunistic bacteria such as Pseudomonas, indicating potential environmental stress or microbiome disruption.
Antibiotic resistance profiles mirror human impacts
The bee microbiome carried 173 antibiotic resistance genes, including multidrug-resistant types that varied widely between sites. Although overall ARG levels were low, their distribution suggests exposure to different microbial communities or pollutants across the city.
“Wild bees silently accumulate signals of ecological stress from limited floral resources to traces of antibiotic resistance,” said Dr. Tang. “These microbial cues can help identify threats to both pollinators and urban ecosystems.”
A diverse virome reveals pathogen spillover
The bees’ gut virome contained a broad array of previously unknown bacteriophages as well as the Apis mellifera filamentous virus (AmFV), a major honeybee pathogen. Its presence at multiple sites suggests a potential virus transmission via shared floral resources between managed honeybees and wild species.
Network analyses showed that phages played a key role in stabilizing gut microbial communities, while shifts in viral composition corresponded to disruptions in the bacterial community.
Microbial networks as early-warning indicators
The study found that bee gut ecosystems containing both bacteria and viruses were more resilient than bacteria-only communities. Reduced numbers of lytic phages, combined with increases in opportunistic bacteria and animal viruses, marked sites experiencing potential environmental stress.
“While our work focuses on a single bee species in one city, the approach is widely scalable,” Dr. Tang added. “We hope these methods will inform pollinator-friendly urban planning and help develop early-warning microbiome biomarkers aligned with One Health principles.”
Conclusion:
Urbanization fragments habitats, alters plant diversity, and exposes wildlife to pollutants and pathogens. Traditional biodiversity surveys rarely capture the physiological stress or microbe-mediated challenges faced by species such as wild bees, which play vital roles in pollinating urban and agricultural plants.
Metagenomic sequencing, analyzing all DNA within an organism’s gut, provides a window into nutrition, microbial symbiosis, pathogen exposure, and environmental contaminants. This study demonstrates that such data can serve as a sensitive measure of wildlife well-being in human-dominated landscapes.
About the research team:
The study was led by Dr. Min Tang, Department of Biosciences and Bioinformatics at XJTLU. Her research group focuses on the health of wild bees, host–symbiont interactions, conservation biology, and microbiome ecology. Integrating metagenomics, computational biology, and environmental science, the team investigates how insects respond and adapt to rapidly changing environments. The group brings together biologists, environmental scientists, and bioinformaticians, reflecting XJTLU’s interdisciplinary research culture.
Read the full article here: https://onlinelibrary.wiley.com/doi/10.1111/1744-7917.70051
Andrew Smith
Charlesworth Publishing Limited
+ +44 7753 374162
marketing@charlesworth-group.com
Visit us on social media:
LinkedIn
YouTube
Other
Legal Disclaimer:
EIN Presswire provides this news content "as is" without warranty of any kind. We do not accept any responsibility or liability for the accuracy, content, images, videos, licenses, completeness, legality, or reliability of the information contained in this article. If you have any complaints or copyright issues related to this article, kindly contact the author above.